Our research interests focus on developing engineering tools to solve biological problems. Our goal is to bridge the gap between biological and engineering paradigms. In particular, we are interested in:
- Mesenchymal stem cell differentiation for bone tissue engineering
- Developing biosensing tools for biomarker detection, single cell analysis, and disease diagnostics
- Developing microfluidic devices to study cell migration during cancer metastasis
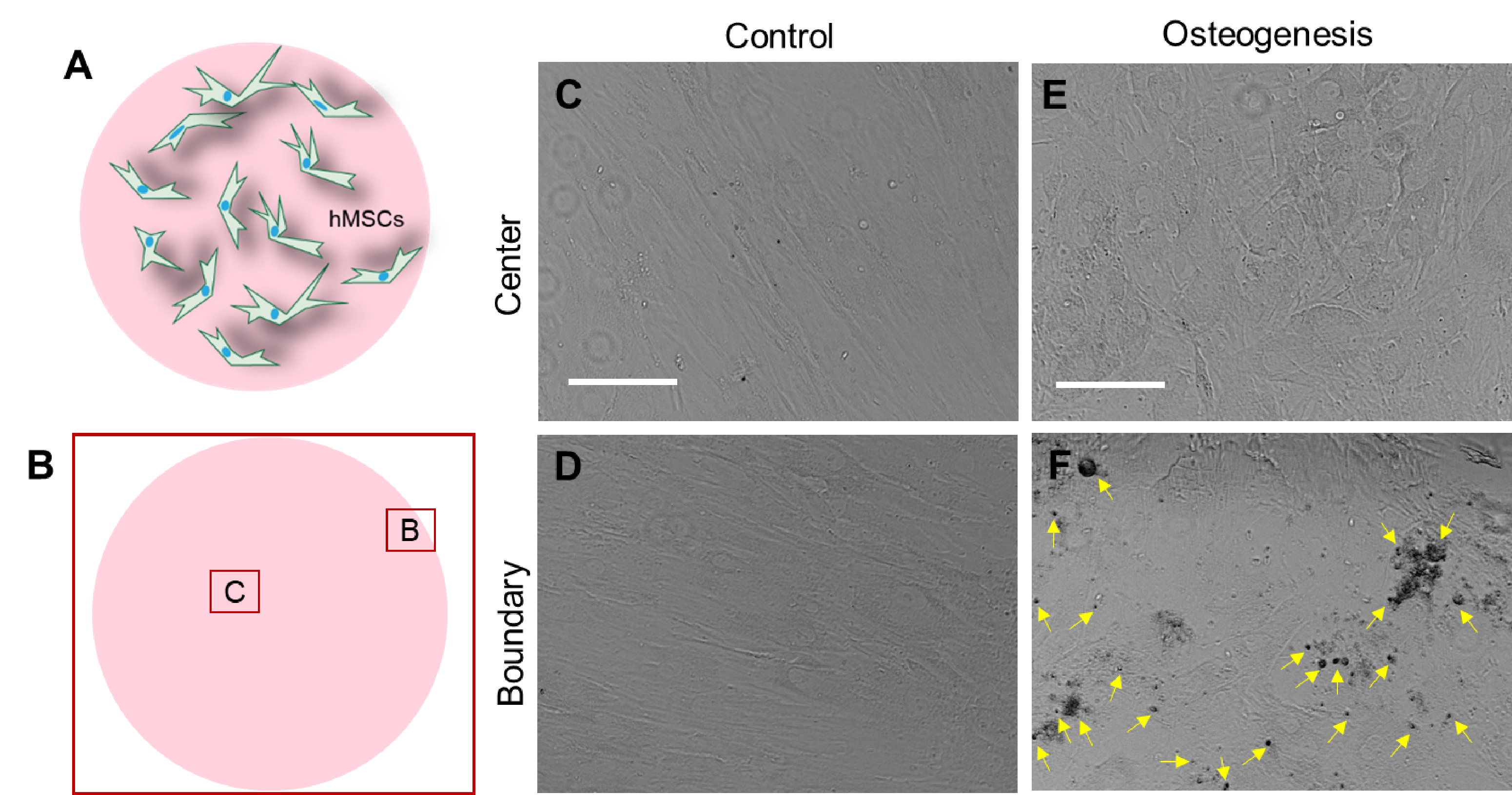
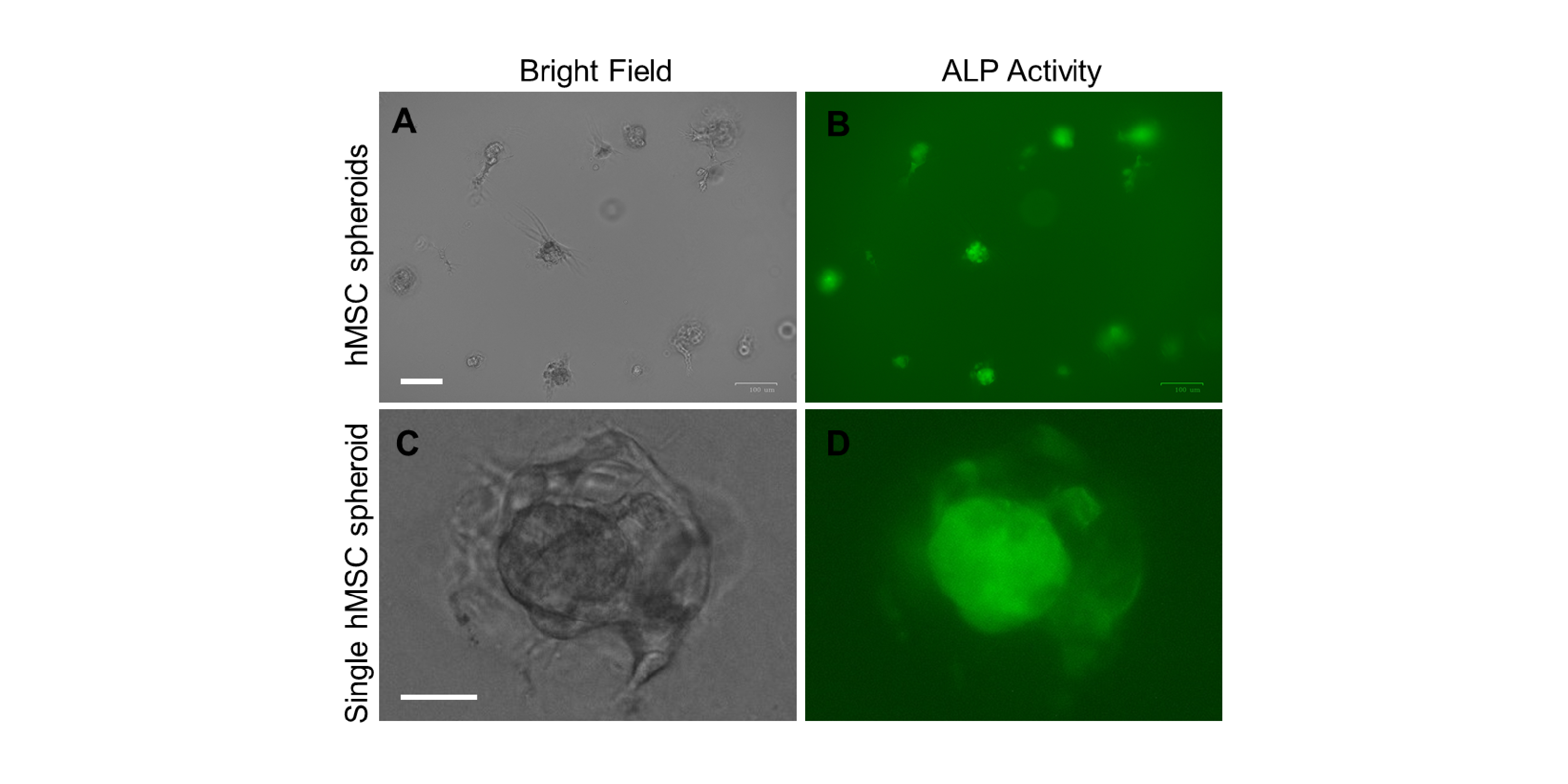
Human Mesenchymal Stem Cells (hMSCs) Differentiation
In bone tissue engineering, a critical factor for bone tissue repair and regeneration is the usage of appropriate cells. Human mesenchymal stem cells (hMSCs) are an attractive cell type due to their potentials of extensive expansion and osteogenic differentiation. hMSCs are multipotent cells which have abilities to differentiate into osteoblasts (bone cells), chondrocytes (cartilage cells), adipocytes (fat cells) as well as other connective tissues. Although hMSCs have been used for bone tissue engineering for decades, it is still challenging to achieve successful and complete osteogenic differentiation.
Recently, there is an increased tanet that mechanical force plays a critical role in the regulation of stem cell differentiation. Stem cells actively sense, adapt and respond to their surrounding microenvironment and interactively responding to external signals.
This project aims to mimic in vivo bone marrow microenvironment for the study of mesenchymal stem cells differentiation. This engineered microenvironment will enable the investigation of hMSC differentiation in a 3D tissue-like environment. The project will uncover the mechanisms underlying physical and biochemical regulation of hMSCs differentiation, which will have a profound impact on the bone tissue engineering and bone regeneration.
Single Cell Biosensors for Multiplex Gene Detection
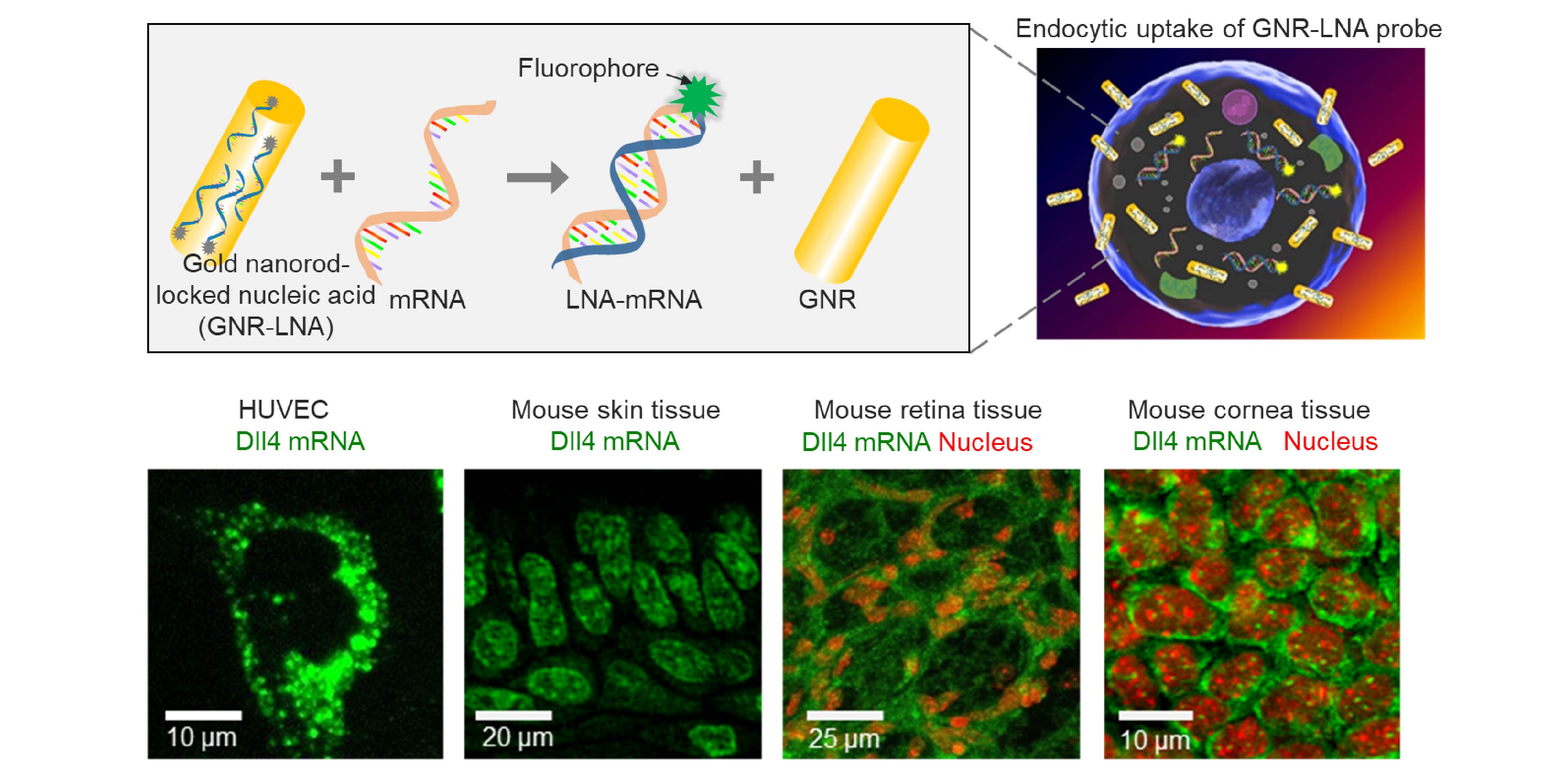
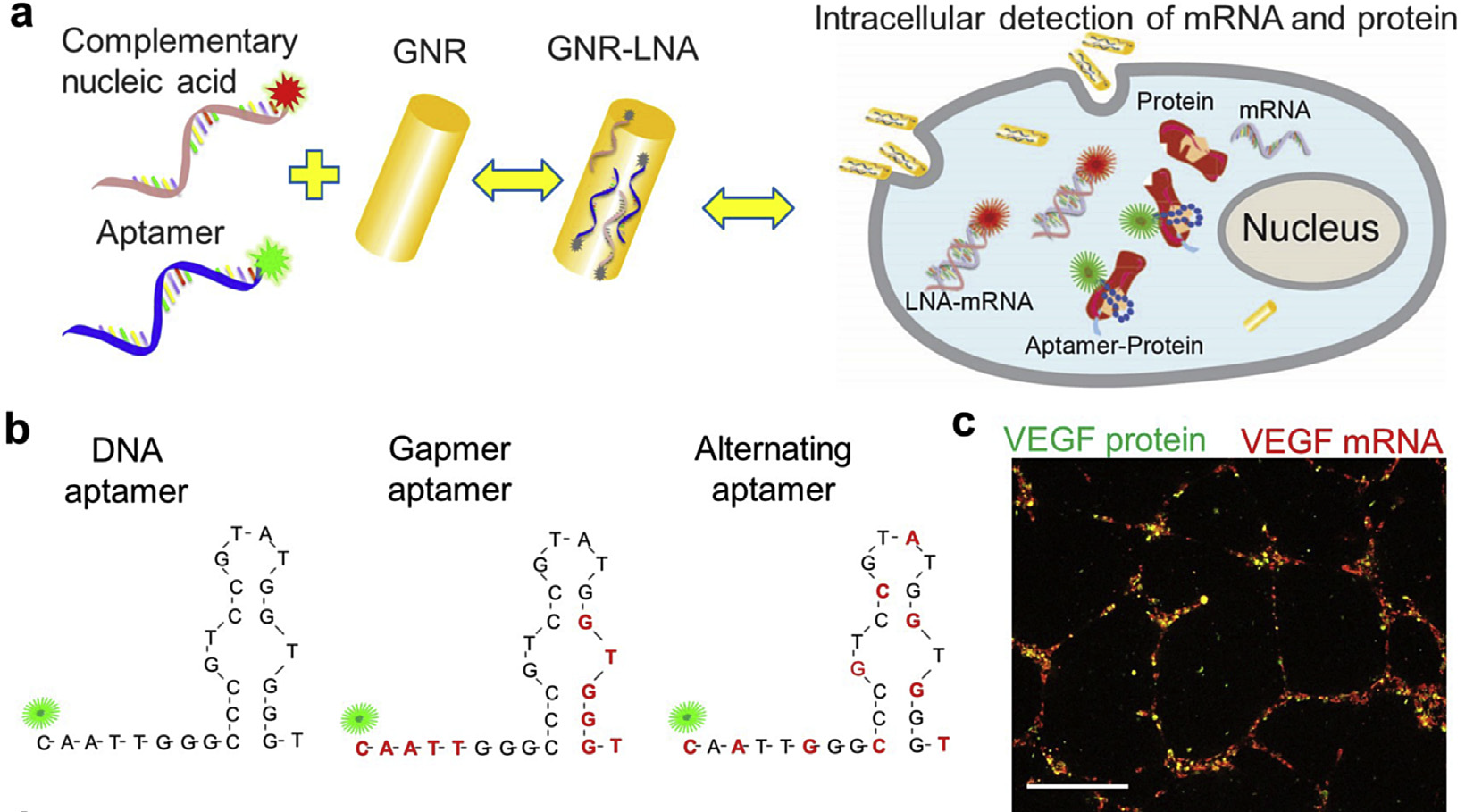
In biological systems, one fascinating and intriguing question is how individual cell senses and respond to local changes to accomplish self-organization and self-healing during both tissue and organ development and repair. During this complex biological process, cells sense internal and external forces which give rise to genetic signaling for self-organization. Understanding the cellular and molecular mechanisms during this dynamic morphogenic and regenerative processes in biological tissues will have important implications in development and regenerative medicine. Nevertheless, the elucidation of the cellular self-organization processes is often hindered by a lack of effective tools for detecting and monitoring the spatiotemporal gene expression distribution and perturbing the cell self-organization processes in living cells and tissues.
To address this critical challenge in monitoring spatiotemporal gene expressions, we developed a multimodal gold nanorod-locked nucleic acid (GNR-LNA) composite for single-cell gene expression analysis in living cells and tissues at both transcriptional and translational levels, Using this GNR-LNA probe, we have demonstrated that this nano-biosensor can detect intracellular mRNA and protein expression in living cells, capillary networks, as wells as mice lung, retina and liver tissues. The ease of this nano-biosensor also enables real-time detection of other biomarkers like microRNA (miRNA), noncoding RNA, mRNA and proteins separately and simultaneously.
Reference:
- S. Wang, Y. Xiao, D. D. Zhang, and P. K. Wong. A gapmer aptamer nanobiosensor for real-time monitoring of transcription and translation in single cells, Biomaterials, Vol. 156, pp. 56-64, 2018. (PDF)
- Y. Zheng*, S. Wang*, X. Xue, A. Xu, W. Liao, A. Deng, A. Liu, and J. Fu. Notch signaling in regulating angiogenesis in a 3D biomimetic environment, Lab on a Chip. Vol. 17, pp. 1948-1959, 2017. (Featured as back cover article, * equal contribution) (PDF)
- S. Wang, J. Sun, D. D. Zhang, and P. K. Wong, A nanobiosensor for dynamic single cell analysis during microvascular self-organization, Nanoscale. Vol. 8 (38), pp.16894-16901, 2016. (Featured as cover article) (PDF)
- S. Wang, R. Riahi, D.D. Zhang, and P. K. Wong. Single cell nanobiosensor for dynamic gene expression profiling in native tissue microenvironments, Advanced Materials. Vol. 27, pp. 6034-6038, 2015.(Featured as cover article) (PDF)
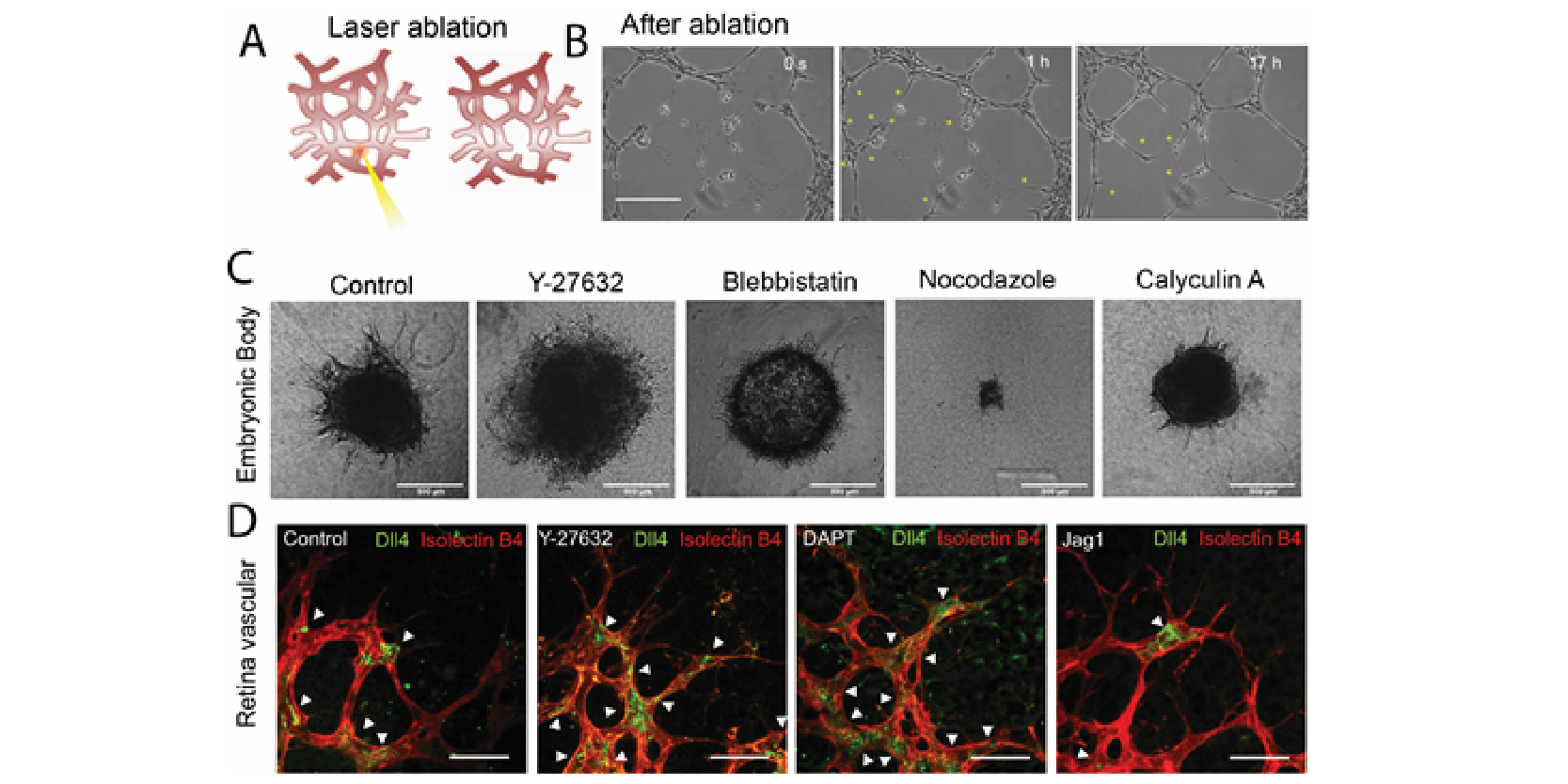
- R. Riahi, J. Sun, S. Wang, M. Long, D.D. Zhang, and P. K. Wong. Dll4 signaling and mechanical force regulate leader cell formation during collective cell migration, Nature Communications. Vol.6, 6556, 2015. (PDF)
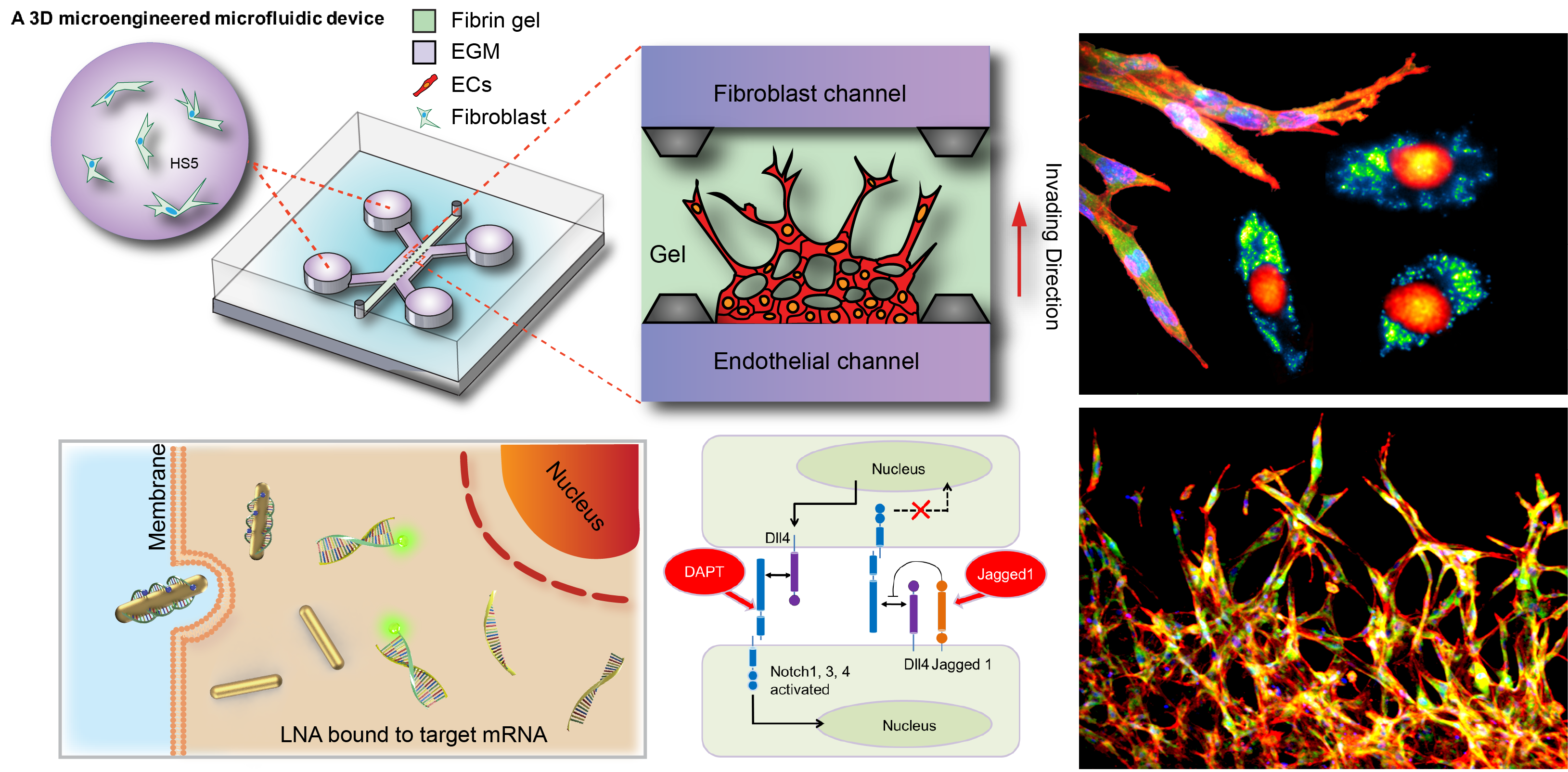
Mechanoregulation of Tissue Morphogenesis and Angiogenesis
Regenerative medicine holds great promise in treating degenerative diseases by stimulating damaged tissue to repair themselves, or replacing them with engineered tissues when the body cannot heal itself. A fundamental understanding of the cellular and molecular regulatory mechanism in tissue regeneration and the ability to modulate these processes are critical to the ultimate success of regenerative medicine.
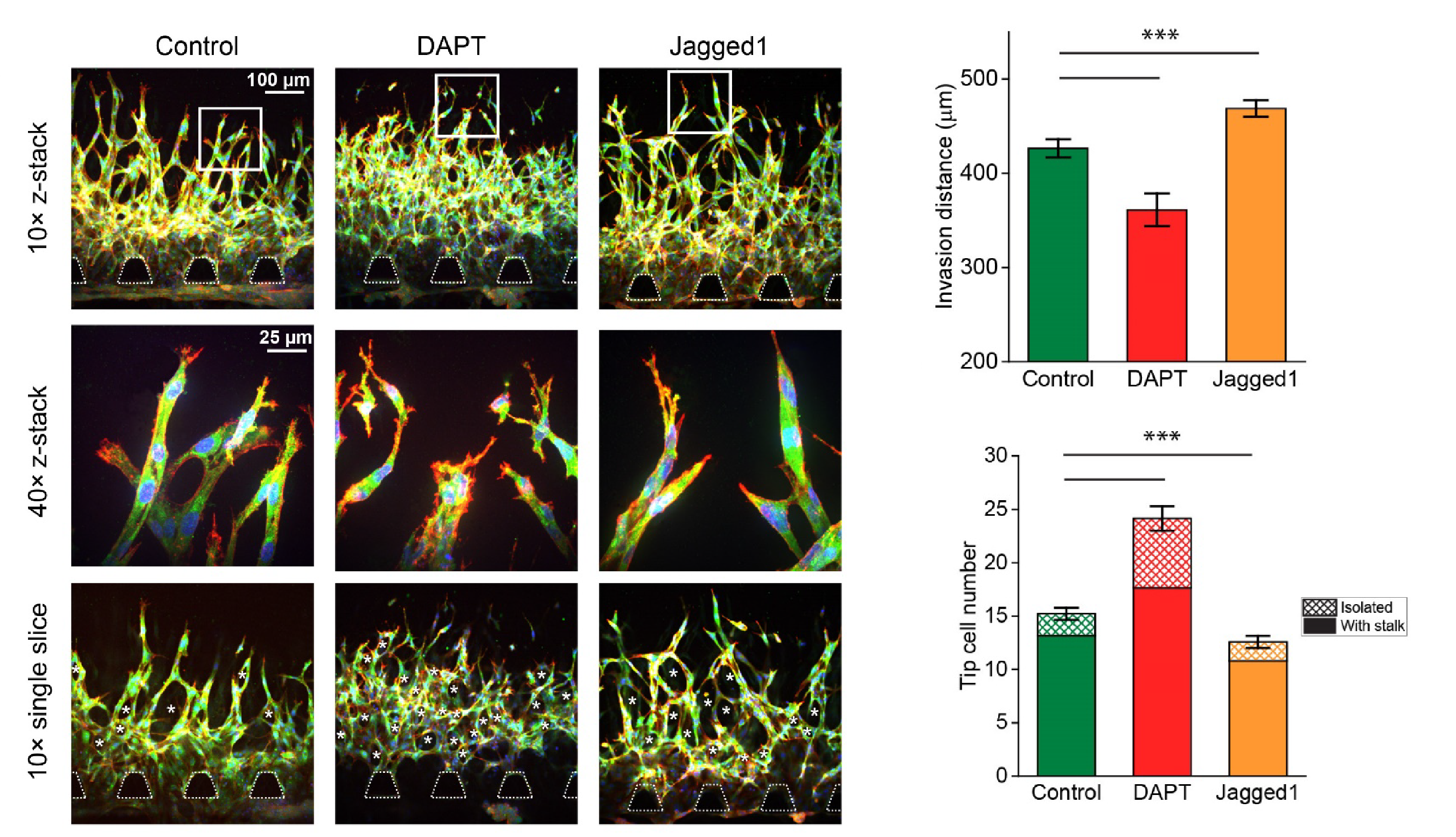
Recently, it has been recognized that mechanical forces play important roles in vascular development during tissue growth and regeneration. Nevertheless, the process by which mechanical force controls the vascular architecture remains poorly understood. Here, we have demonstrated that mechanical force negatively regulates tip cell formation via Notch1-Dll4 signaling in mouse retinal angiogenesis in vivo, sprouting embryoid bodies, and human endothelial cell networks in vitro.
Taken together, our results suggest mechanical force regulates the vascular architecture by modulating angiogenic sprouting via Notch1-Dll4 signaling during tissue morphogenesis. The finding will have great significance for various biomedical applications, such as tissue engineering, cancer, and drug screening.
Reference:
- Y. Zheng*, S. Wang*, X. Xue, A. Xu, W. Liao, A. Deng, A. Liu, and J. Fu. Notch signaling in regulating angiogenesis in a 3D biomimetic environment, Lab on a Chip. Vol. 17, pp. 1948-1959, 2017. (Featured as back cover article, * equal contribution) (PDF)
- S. Wang*, J. Sun*, Y. Xiao*, Y. Lu, D. D. Zhang, and P. K. Wong. Intercellular tension negatively regulates angiogenic sprouting of endothelial tip cells via Notch1-Dll4 signaling, Advanced Biosystems. Vol. 1, 1600019, 2017. (PDF)
Droplet-based Microfluidics
Microfluidic techniques have been widely developed and applied to different fields including single-cell analysis, cancer metastasis and angiogenesis over the last two decades. However, microfluidic encapsulation of bacterial or mammalian CFE systems has only been studied in recent years. The microfluidic platform offers an efficient approach for high encapsulation rate with controllable compartment sizes. In our study, LNA probe in conjunction with a fluorescent reporter protein, and HeLa-based CFE were encapsulated in single-emulsion droplets using a microfluidic device.
The microfluidic device is robust, easy to implement and could be used repeatedly. Waterin-oil droplet microfluidics provide an effective method to efficiently reduce sample volume to picolitre or femtolitre range, compared to the bulk reaction volume of microliters. By encapsulating HeLa based CFE in picoliter-sized droplets, the TX–TL dynamics were visualized and monitored in the cell-sized droplets.
Reference:
- S. Wang, N. Emery, A. Liu. Simultaneous monitoring of transcription and translation in mammalian cell-free expression in bulk and in cell-sized droplets, Synthetic Biology, Vol. 3 (1), pp. 1-9, 2018. (PDF)
- Y. Zheng*, S. Wang*, X. Xue, A. Xu, W. Liao, A. Deng, A. Liu, and J. Fu. Notch signaling in regulating angiogenesis in a 3D biomimetic environment, Lab on a Chip. Vol. 17, pp. 1948-1959, 2017. (Featured as back cover article, * equal contribution) (PDF)